Although that’s kind of what I felt like telling him, unlike HAL-9000, I do still need Dave. So I finally buckled down this morning to step through the rest of his manuscript, hunting down typos and noting what constructive things I can say about each section.
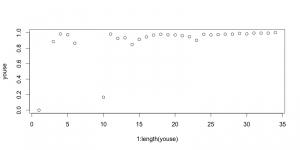
I had left off the last time at the section “Preservation at the species level”. In his table of % preservation of extant species, he gives diatoms only a <50% percent score, but gives the total extant diversity as 1,500. While I have no doubt that there are at least that many diatom species today, I’m not convinced that there are anywhere near that many that are commonly found as pelagic, planktonic species. Much of the extant diatom diversity (which far exceeds 1,500) is terrestrial/lacustrine or benthic/epiphytic marine, and is sampled in the Souria survey as tychoplankton. I chased up the Souria, 1991 species to see where that number comes from. Indeed, between 37 and 44% (depending on whether you choose the low or high end of their range estimate) of the diatom diversity reported consists of pennates—which are mostly benthic. I think if you were to include only those species that are, like radiolarians, exclusively marine and planktonic, the % of preservation would be far higher. It would also be nice to have the number of preserved fossil species in the table (not just the %age), and a source reference for that number, cited in the table legend. Well, I just checked the Kooistra chapter in Falkowski & Knoll, and he cites numbers between 5,000 and 10,000 for the diversity of strictly marine planktonic diatoms. Oops. Well, scratch that, then.
Much of the rest of the section is OK, although there’s a paragraph on page 7 I have a bit of a hard time with. I don’t think I’ll comment on it, but Dave makes the point that even though preservation is so good, it’s possible that there were times when very ill-preservable species of radiolarians evolved, that left no record. Sure, it’s possible, but so what—it’s also possible that the Earth was invaded time and again over its history by purple alien cloud-people who left no footprints and no traces of their influence. Sure, it’s possible.
He also makes the point that there are ‘significant gaps’ because some regions, such as the gyres, don’t leave much of a microfossil record at all. This is true, but again it seems like peanuts compared with the sort of comparatively appalling preservation characteristic of invertebrate fossils on the shelves.
When Dave talks about hiatuses, again, he makes the deep sea record seem worse in comparison to the shelf record, in a way I’m not sure is fair—because he suggests there are no changes in lithology to suggest a hiatus has taken place. My sense is that changes in lithology don’t necessarily help you in shelf sections—sometimes they might represent very little time, and sometimes there can be consistent lithology over many millions of years. I’m not sure the problems of recognizing the partitioning of time in rocks are really meaningfully different in deep and shallow sediments.
Fortunately Dave ends the section on quite a positive note; perhaps I didn’t quite appreciate this on first reading. He says that in spite of all of this, the record is really good—species level evolution for entire clades for most of the biogeographic provinces over the past 100 million years.
Unfortunately, he then launches into a diatribe about how poorly this record is recovered and documented. This is the next section, “Recovery of Deep-sea Fossil Material”. This section begins with a description of piston-coring, and its fantastic coverage, but the admittedly damning limitation of short timescale. Moving on to deep-sea drilling material, he duly acknowledges the staggering number of fossils already available for study (at least 10^15 specimens, a million times more than all the world’s natural history museums combined), and the fact that most of these come with coeval paleoenvironmental data. Surprisingly (for me, given what I remembered from my first reading of the paper) he also ends this section on a positive note, namely that the record is nearly complete at the species level, given that the MRC holds more than 100 samples per million years for most of the Cenozoic.
In the next section, “IRAT—Imperfections in the Existing Dataset”, he explains why the data generated from these samples is less than complete, and why it’s a problem to use them for paleobio research. “Incomplete Data” outlines a problem with how species are recorded on a slide. Rather than the ideal model, in which the paleontologist records the taxonomic identity of a certain number of specimens, and then moves on to the next slide, thereby obtaining a random and unbiased subsample of the sample in hand, the situation is usually as follows. The paleontologist has a list of taxa that is as short as it can usefully be, and he records presence/absence (or abundance) of those taxa in order to determine the age of the sample.
But on top of this, the paleontologist often records some additional taxa, which do conform to the random sampling. Crucially, he states that “the differences in the average reported diversity per sample/study simply reflect the average practical size of a taxonomic list, and do not have a necessary relationship to actual real sample diversity”. Now this seems to be the key sentence. Does this mean each study has a different taxonomic list, and that’s what determines list length, more so than underlying diversity? If so, this should be easy to test (and I think Dave should do this if he wants to back up his argument): what publication a sample is from should be a better predictor of list length than what time it’s from. So, if the Neptune database has publication information (which I hope it does), you should be able to parse the data by time bins vs. by publication, and see if the variability is better described by what time bin samples are in, or by what publication they’re from. This could be compared by Akaike weights, for example.
The next paragraph—on page 11—is really quite confusing, and stands at the heart of the part of this paper that affects what I’ve been thinking about and doing with diatom diversity. Dave states that most data is collected by his “model C”, so the model where paleontologists record the presence/absence of taxa on a list, plus whatever other taxa they fancy. He states that this leads to a correlation between sample availability and total diversity, but not because of the reason we might think (i.e., going up a collector curve)—but rather, because sample availability is correlated with taxonomic effort. I think what he means here is that sections with more samples available have “model B” taxonomic lists that are longer than sections with few samples. It seems to me, though, that this reduces down to the same thing as collector curves, albeit via the detour of constructing a list: the more diverse-seeming assemblages seem thus because they have longer “model B” taxonomic lists, not because they’ve had more random samples taken, but the reason they have longer taxonomic lists is because there is more “sample availability”, as Dave puts it, which I think means… they have been more extensively—randomly—sampled.
In the next paragraph, he rallies support from a figure (figure 8) that I just don’t understand. The point he’s trying to make is that species are more rarely reported than they should be, I think; what he shows is a histogram of the number of samples from which a radiolarian taxon is reported; 100+ taxa show up in only 1-5 samples, and 40+% of taxa show up in 25 samples or fewer. Besides the fact that the plot is confusing (not clear what the inset plot is, vs. the main plot, nor what the total number of samples is) and the calculation in the figure caption is impossible to follow, I’m not sure this addresses the same point as the preceding paragraph. That paragraph was trying to say that subsampling exercises wouldn’t work because many, or most, of the taxa in the database will be from “model B” lists of stratigraphic marker species. Apart from the fact that this might not be true (see below), the point explored with figure 8 is different.
Is it true that “model C” makes subsampling impossible? I think Dave might have his answer backwards, actually. If the “model B” list is consistent over time (and I’m not sure what Dave’s stance is on that—he seems to want it both ways at the beginning of this section), then you might actually be making a much fairer comparison if you are subsampling by lists, because each list you pull will be comparing apples to apples in its “model B” component. In addition it provides its “model A” component, but that should be subject to the same qualifying properties of random sampling as sanctioned by Dave in the beginning of the section, so it should behave well under subsampling. So, aren’t we actually improving things in this way?
Of course, if lists are different depending on what time interval we’re looking at, then I think the “model C” argument just breaks down to a “model A” scenario, more or less.
The main point here, though, is that this does not distinguish the microfossil record in any way from the rest of the fossil record—dominated by shelf invertebrates—as recorded in PBDB. That record is also a combination of biostratigraphic occurrence data of a limited, and commonly represented, stratigraphically informative species, and a more or less random sampling of other taxa. How does that make the microfossil any worse?
Moving on, the section “Reworking” opens with the claim that reworking affects only the microfossil record, a claim I think can hardly be considered true. I don’t have any great references at hand, but can offer one (sight unseen, thanks to a lapsed subscription to Lethaia, cheers Harvard): Fürsich, F.T. 1978. The influence of faunal condensation and mixing on the preservation of fossil benthic communities. Lethaia, Volume 11, Issue 3, pages 243–250. Also Kidwell, S.M., 1998, Time-averaging in the marine fossil record: overview of strategies and uncertainties: Geobios, v. 30, p. 977– 995. Kidwell, S.M., and Bosence, D.W.J., 1991, Taphonomy and time averaging of marine shelly faunas: in Allison, P.A., and Briggs, D.E.G., eds., Taphonomy: Releasing the Data Locked in the Fossil Record: Plenum Press, New York, p. 115–209. Recent study: DeFrancesco, C. and Hassan, G.S. 2008. PALAIOS; v. 23; no. 1; p. 14-23.
And again, these issues are all the same issues that befall the macrofossil record, too—I don’t think an obviously reworked specimen will be reported by a trilobite worker as occurring in the formation in which it was found.
The first sentence of the “Age Model Problems” sentence really says it all—they’re way better for the marine microfossil record than for any other record we have. And that should be the focus of the paper, not all the things that are wrong with it! In diversity studies, a 1-my error is not a problem if we use 2-my bins. Also, this error is unbiased—and this is a critical point—so for macroevolutionary studies, it really shouldn’t matter. As long as it affects everything equally, and more or less evenly throughout time, we should be golden as long as the signal we’re trying to see is strong enough.
The same “but it’s even worse in the rest of the fossil record” argument can be brought against the “Taxonomy” section, which says because there’s convergence and some morphospecies overlap through time, but this has got to be a pretty minor problem and should only cause ranges to extend very slightly.
“Reworking, Age Model Errors and Macroevolutionary Metrics”. Dave shows his calculations (or their result) that suggest 5% of radiolarian LADs in Neptune are off, and 3% of FADs. He adds it up to a total error of 8% of all occurrences being outside the true range of the species. This leaves the apparent ranges of many taxa extended beyond their true ranges, which is a big difference to most of the rest of paleontology, where the opposite is the case (this is actually a really good point, I think). But because taxa are rare, Dave proposes using range-through; the problem then becomes the artificial range extension and how to deal with it.
I think it might be worth piping up here and putting in a word for Alroy and his distaste for range-through, because of the ugly edge effects it causes. But to illustrate the downfalls of range-through, Dave does something really sketchy here that bothers me a lot. He takes a 1 my time bin for forams in Neptune, and compares what’s found there to what’s supposed to be there based on the biostratigraphic framework.