March Madness Day 19: No Energy
ø
Motivation is at a new low. I just can’t seem to summon the energy to do any work. I am just dispirited.
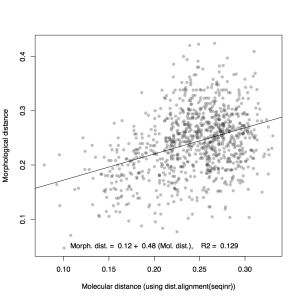
It took me a long time to get going, once again, and I spent the morning catching up on DSA posts for last week. Scott Edwards got back to me by email and I tracked him down in the afternoon for a chat about molecular vs. morphological distances. He was incredibly nice—just a really friendly guy—and I did get one or two references out of him. But in general I found it quite hard to get much out of him that was directly helpful. What I had been hoping for was context and explanation of the plots I’d made, basically a sense of what that distance comparison would look like for other groups of organisms, and whether my plot is expected or unexpected. Instead, I found that he seemed most interested in advising me to do other analyses with the data I have—mainly studying the evolution of characters on the tree, relating morphological change to speciation events, for example, seeing if disparity grows anagenetically or cladogenetically, and so forth. These are of course really interesting questions, but they’re a lot of extra work and analysis that I don’t think I have to the time to do. There just seemed to be an almost unbridgeable gulf between my approach (focused on morphology alone, making comparisons to phylogeny and diversity) and his approach, which appeared to be entirely phylogenetically focused.